You have read the basics of nucleic acids in my previous post. Let’s have a deeper look at nucleotides in this post.
The nucleotide sequence of DNA dictates the amino acid sequence of a protein and nucleotide sequence of RNA.
As seen before, a nucleotide has three components: a nitrogenous base, a pentose sugar and a phosphate attached to the sugar. A nucleotide without phosphate is a nucleoside.
Bases:
The nitrogen containing bases originate from purines and pyrimidines, the two parent compounds.
The 5 major bases fall into two categories based on their parent compound-
Purines give rise to two bases: Adenine (A) and Guanine (G)
Pyrimidines give rise to three bases: Thymine (T), Cytosine (C), and Uracil (U)
Purines and pyrimidines are aromatic and are weakly basic and are hence bases. The aromatic nature of these purines and pyrimidines is important for its electron distribution and the light absorption of nucleic acids. In fact, the electron delocalisation is what gives most bonds the partial double bond character.
These bases are hydrophobic at neutral pH.
Pentose Sugar:
They fall into two categories:
Ribose found in Ribo Nucleic Acid (RNA) and
Deoxyribose found in Deoxyribo Nucleic Acid (DNA)
Nucleotides
They contain three components:
- A nitrogenous base
- A pentose sugar
- A phosphate group
Nucleosides are nucleotides without a phosphate group.
Based on the pentose sugar, nucleotides are classified into deoxyribonucleotides and ribonucleotides. They are the structural units of DNA and RNA respectively.
Deoxyribonucleotides are further classified as Deoxyadenosine, Deoxyguanosine, Deoxycytosine and Deoxythiamine.
Ribonucleotides are classified as adenosine, guanosine, cytidine and uridine
In addition to the four bases, both DNA and RNA contain some minor bases. Let’s look at some examples:
- Methylated bases in some bacteria or hydroxylated and glycosylated bases in certain viruses
- Some nucleotides also have their phosphate groups attached to positions other that 5’. Eg: adenosine 3’,5’-cyclic monophosphate (cAMP) and guanosine 3’,5’-cyclic monophosphate (cGMP) are important secondary messengers
Linking between nucleotides in a polynucleotide
Polynucleotides as the name suggests are polymers of nucleotides. Eg include DNA and RNA.
The nucleotides within these polynucleotides link to each other through the phosphate group bridges called phosphodiester linkage formed by the joining of 5’-phosphate group and 3’-hydroxyl group.
A short nucleic acid consisting of around 50 or fewer nucleotides are oligonucleotides. Anything longer than that is polynucleotide.
Biosynthesis of purines and pyrimidines nucleotides
Purine nucleotides synthesis
The two purines are Adenine (A) and Guanine (G). So, their parent compounds are
Adenine 5’-monophosphate (AMP/ adenylate) and
Guanine 5’-monophosphate (GMP/ guanylate)
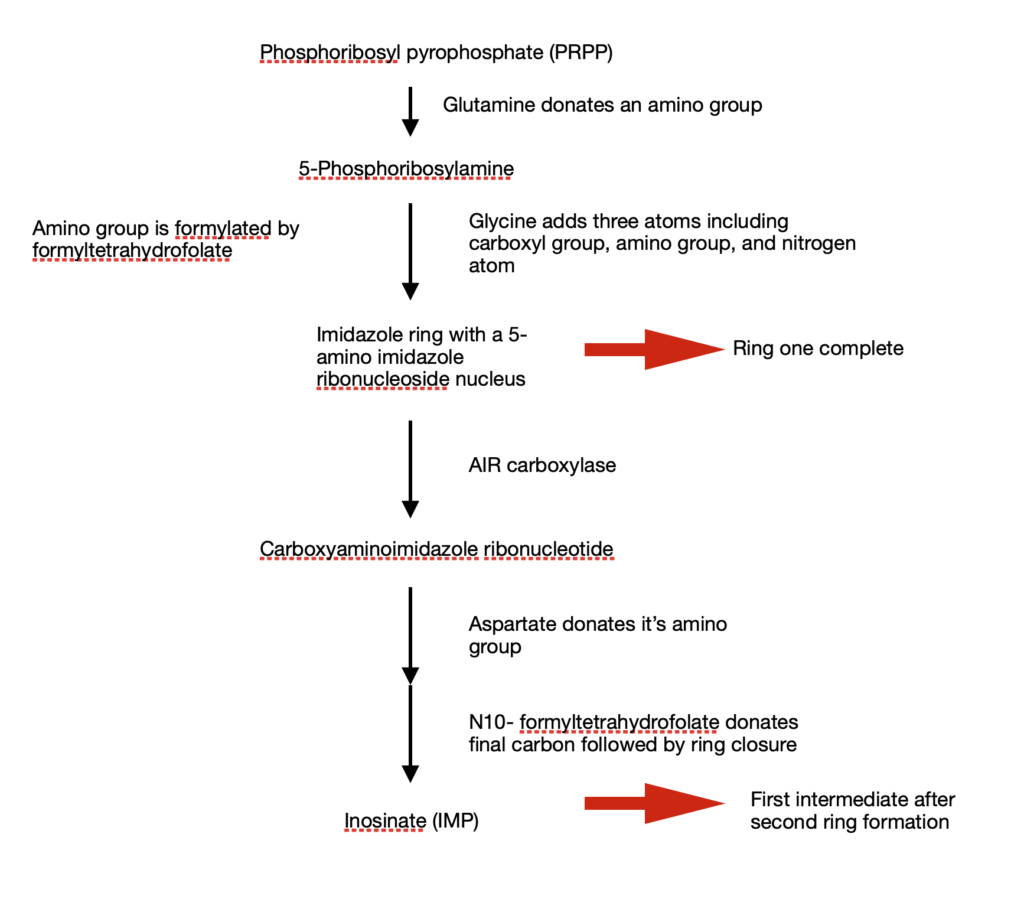
The entire process of purine biosynthesis takes place in 11 steps.
- Glutamine donates an amino group to the parent compound. This amino group attaches to the C1 of phosphoribosyl pyrophosphate (PRPP) to give a highly unstable 5-phosphoribosylamine
- Three atoms from glycine is added to 5-phosphoribosylamine. This condensation requires the consumption of an ATP. Activating the carboxyl group of glycine requires ATP
- After condensation, the amino group of glycine is formylated by N10-formyltetrehydrofolate to give nitrogen
- Glycine contributes a nitrogen group
- Dehydration and ring closure to give the five membered imidazole ring of the purine nucleus. This nucleus ring is called 5 amino imidazole ribonucleoside.
By this step, I ring of the purine structure is complete - 3 out of 6 atoms required for the second ring are in place. A second carboxyl group is added to complete the process. This carboxylation is unique as it uses bicarbonate instead of biotin
- Rearrangement: carboxylate is transferred from amino acid group to position 4 of the imidazole ring
Steps 6 and 7 take place only in bacteria and fungi
In humans, 5 aminoimidazole ribonucleotide is directly carboxylated to carboxyaminoimidazole ribonucleotide by AIR carboxylase
In steps 8 and 9, aspartate donates it’s amino group - Formation of amide bond
- Elimination of carbon skelton of aspartate
- Final carbon is donated by N10- formyl tetrahydrofolate
- Second ring closure resulting in a fused ring structure of purine nucleus
Step 1 to 11 will finally result in an intermediate (with a full purine nucleus) – Inosinate (IMP)
In short,
Pyrimidine nucleotides synthesis
Pyrimidines are cytosine and uracil. So, their parent compounds are cytidine 5-monophosphate (5CMP and cytidilate) and uridine 5-monophosphate (5UMP and uridylate) respectively.
Six membered ring is formed and then attached to ribose 5-phosphate. The attachment requires carbonyl phosphate. To make carbonyl phosphate (for use in pyrimidine synthesis), we require carbamoyl phosphate synthetase II (enzyme 1).
- Carbamoyl phosphate + aspartate—aspartate transcarboxylase (enzyme 2)—— N-carbamoyl aspartate
- N carbamoyl aspartate——-removal of water using dihydroorotase (enzyme 3)——— L-dihydroorotate
This step ensures the closure of the pyrimidine ring - L-dihydroorate—-oxidation—— orotate (NAD+ is the electron donor)
In humans, these three enzymes are a part off a single enzyme- CAD - Ribose 5-phosphate chain of PRPP + orotate——-orotidylate
- Orotidylate——-decarboxylated——- uridylate
- Urildylate—-phosphorylated——— UTP
- UTP——cytidylate synthase———- CTP
nitrogen donor is either glutamine or NH4+
In short,
| Step | Substrate | Enzyme | Product |
| 1 | Carbamoyl phosphate synthase II | Carbamoyl phosphate | |
| 2 | Carbamoyl phosphate + aspartate | Aspartate transcarboxylase | N-carbamoyl aspartate |
| 3 | N carbamoyl aspartate | Dihydroorotase | L- dihydroorotate |
| 4 | L-dihydroorotate | Oxidation and NAD+ as electron donor | Orotate |
| 5 | Orotate + Ribose 5-phosphate | Orotidylate | |
| 6 | Orotidylate | Decarboxylase | Uridylate |
| 7 | Uridylate | Phosphorylase | UTP |
| 8 | UTP | Cytidylate synthase | CTP |
Degradation of purines and pyrimidines
Purine nucleotides lose their phosphate through 5’-nucleotidase.
Adenine catabolism
Adeninylate———adenosine——deaminates ——-Inosine deamination requires adenosine deaminase
Inosine——hydrolysed—— hypoxanthine and D-ribose
Hypoxanthine—-oxidised——xanthine—-xanthine oxidase——uric acid
So, in short
| Step | Substrate | Process | Enzyme | Product |
| 1 | Adenylate | Adenosine | ||
| 2 | Adenosine | Deaminates | Adenosine deaminase | Inosine |
| 3 | Inosine | Hydrolysed | Hypoxanthine and D-ribose | |
| 4 | Hypoxanthine | Oxidised | Xanthine | |
| 5 | Xanthine | Oxidised | Xanthine oxidase | Uric acid |
Guanine catabolism
GMP—— hydrolyse—- Guanosine
Guanosine—-cleave—— Guanine
Guanine——removal of amino groups—— Xanthine
Xanthine—- Xanthine oxidase—— Uric acid
So, in short
| Step | Substrate | Process | Product |
| 1 | GMP | Hydrolyse | Guanosine |
| 2 | Guanosine | Cleave | Guanine |
| 3 | Guanine | Removal of amino groups | Xanthine |
| 4 | Xanthine | Oxidation | Uric acid |
Pyrimidines give NH4+ upon degradation which enters urea synthesis
Eg: Thymine——-degrade—- Methylmalonylsemialdehyde
Methylaminosemialdehyde——propionyl CoA and methyl malonyl CoA—- Succinyl CoA
Nucleoside Monophosphate to Nucleoside Triphosphate
Most nucleotides, after synthesis are converted to nucleoside triphosphate. This takes place in 4 steps.
- Phosphorylation of AMP to 2 ADP with the help of adenylate kinase
- Phosphorylation of ADP by glycolytic enzymes or oxidative phosphorylation
- ATP helps in the formation of other nucleoside diphosphate with the help of nucleoside monophosphate
- Nucleoside diphosphate is converted to Nucleoside Triphosphate by nucleoside diphosphate kinase
So, in short
| Step | Substrate | Process | Enzyme | Product |
| 1 | AMP | Phosphorylation | Adenylate kinase | ADP 2 |
| 2 | ADP | Phosphorylation | Glycolytic enzymes | ATP |
| 3 | ATP | Nucleoside monophosphate | Other Nucleoside Diphosphates | |
| 4 | Nucleoside Diphosphate | Phosphorylation | Nucleoside diphosphate kinase | Nucleoside Triphosphate |
Derivation of deoxyribonucleotides
Their derivation is directly from ribonucleotides by direct reduction at 2’ carbon atom of D-ribose to give 2X deoxyderivatives. This reaction is catalysed by the enzyme ribonucleotide reductase.
Denovo and Salvage pathway
Denovo pathway
- It happens through metabolic precursors like amino acids, ribose 5-phosphate, CO2 and NH3
- In Denovo synthesis, nucleotides are not synthesised and then attached to the ribose. They are instead attached to the ribose while being synthesised
- Enzymes are largely multi enzyme complexes
- Important precursor for de novo synthesis of both purines and pyrimidines is Phosphoribosyl pyrophosphate (PRPP)
Salvage pathway
- They recycle the free bases and nucleosides released from nucleic acid breakdown. It is much simpler than the Denovo pathway
Examples of salvage pathway
Free adenine + PRPP—-adenosine phosphoribosyltransferase—— AMP+ PPi
Free guanine and hypoxanthine are salvages in the same way by hypoxanthine guanine phosphoribosyltransferase
Now that you have got an understnanding of Nucleic Acids? Check if you can answer this question.
Question: In a test tube, RNA and DNA are hydrolysed under alkaline conditions. Which of them will hydrolyse rapidly and why?
Comment your answers below.

Very helpful!
Thanks for sharing!